Efficiency
- quickly write, revise, run, and revisit your code
Transparency
- let others see & understand what you're doing
Reproducibility
- enable anyone to run your analyses
10 March, 2016


for(i in seq_len(n)[-1]) {
mu[i,d]<- X[i-1, d]+((GPP[d]/z)*(light[i,d]/(sum(light[,d]))))+ (ER[d]*0.006944/z)-
(K[d]/(600/(1800.6-(temp[i,d]*120.1)+(3.7818*temp[i,d]^2)-(0.047608*temp[i,d]^3)))^
-0.5) * 0.0069444*( ((exp(2.00907 + 3.22014 * (log((298.15-temp[i,d]) / (273.15 +
temp[i,d]))) + 4.0501 * (log((298.15 - temp[i,d]) / (273.15 + temp[i,d]))) ^ 2 +
4.94457 * (log((298.15 - temp[i,d]) / (273.15 + temp[i,d]))) ^ 3 - 0.256847 *
(log((298.15 - temp[i,d]) / (273.15 + temp[i,d]))) ^ 4 + 3.88767 * (log((298.15 -
temp[i,d]) / (273.15 + temp[i,d]))) ^ 5)) * 1.4276 * bp / 760) *satcor-X[i-1,d] )}
# Compute the O2-specific reaeration coefficient for each timestep
K <- convert_k600_to_kGAS(K600.daily, temperature=temp.water, gas="O2") * frac.D
# Dissolved oxygen (DO) at each time i is a function of DO at time i-1 plus
# productivity, respiration, and reaeration
for(i in seq_len(n)[-1]) {
DO.mod[i] <-
DO.mod[i-1] +
GPP[i] +
ER[i] -
K[i] * (DO.sat[i] - DO.mod[i-1])
}
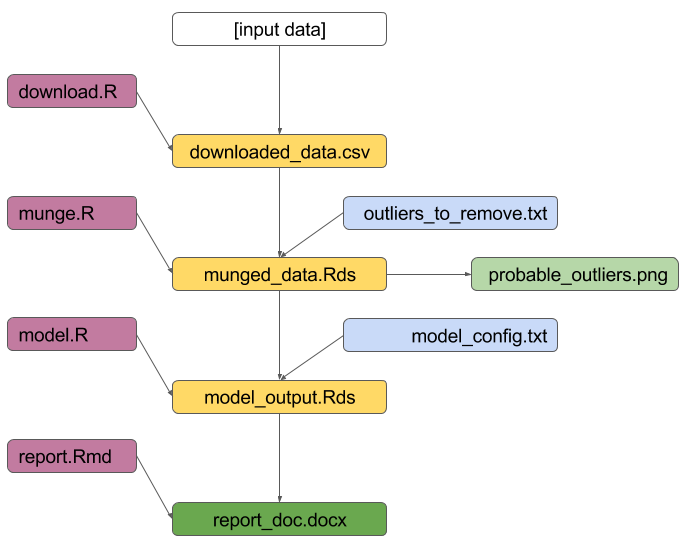
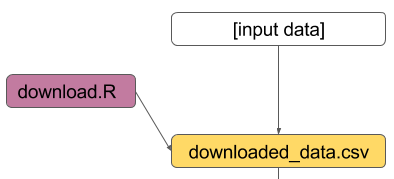
01_download.R
01_downloaded_data.csv
02_munge.R
02_munged_data.Rds
03_model.R
03_model_output.Rds
04_report.Rmd
04_report_doc.docx
+---01_data
| downloaded_data.csv
|
+---02_cache
| a_munged_data.Rds
| b_model_output.Rds
|
+---03_results
| report_doc.docx
|
\---code
01_download.R
02_munge.R
03_model.R
04_report.Rmd
+---01_data | +---code | | download.R | \---out | downloaded_data.csv +---02_munge | +---code | | munge.R | +---doc | | probable_outliers.png | +---in | | outliers_to_remove.txt | \---out | munged_data.Rds +---03_model | +---code | | model.R | +---in | | model_config.txt | \---out | model_output.Rds +---04_report | +---code | | report.Rmd | \---out | report_doc.docx \---lib
+---01_data
| downloaded_data.csv
|
+---02_cache
| a_munged_data.Rds
| b_model_output.Rds
|
+---03_results
| report_doc.docx
|
+---code
| 01_download.R
| 02_munge.R
| 03_model.R
| 04_report.Rmd
|
\---ideas
+---150911_lmer_model.R
| try_mixed_mod.R
|
\---160227_facet_plot.R
plot_a.R
plot_everything.R

download.file() # httphttr::GET() # httpsdataRetrievalgeoknifedataone# Download & unzip the UMESC fisheries data url <- 'http://www.umesc.usgs.gov/data_library/fisheries/LTRM_FISH_DATA_ENTIRE.zip' fishzipfile <- tempfile() download.file(url, destfile=fishzipfile) unzip(fishzipfile, exdir='data', junkpaths=TRUE)
nrow(), length(unique()))sd(), sensorQC::flag())table())lubridate::tz())plot(x=lon, y=lat))
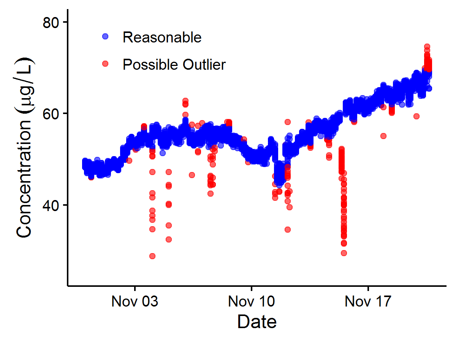



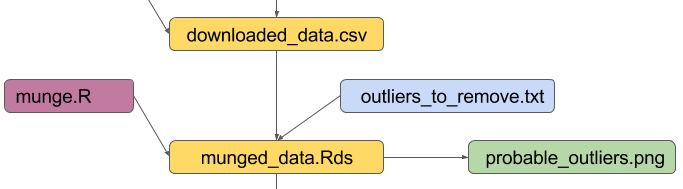
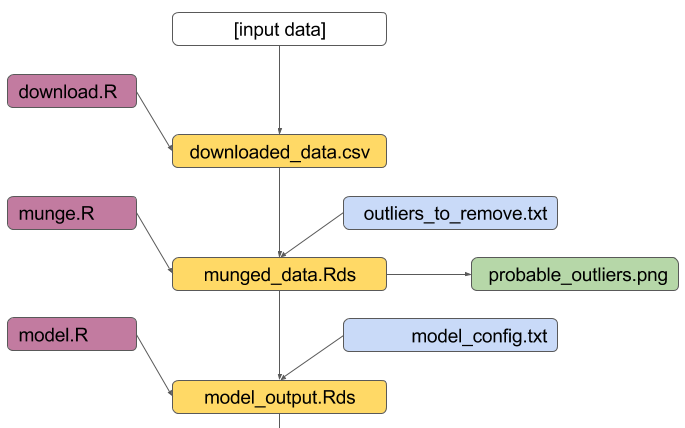
maketarget: dependencies
[tab] system command
01_downloaded_data.csv: 01_download.R
$(R_CMD) 01_download.R`
VAR = value
R_CMD = R CMD BATCH --no-save --no-restore --slave --no-timing
# Macros
R_CMD = R CMD BATCH --no-save --no-restore --slave --no-timing
# Targets
all: 01_downloaded_data.csv
01_downloaded_data.csv: 01_download.R makefile
$(R_CMD) 01_download.R

# Macros
R_CMD = R CMD BATCH --no-save --no-restore --slave --no-timing
# Targets
all: 03_model_output.Rds
01_downloaded_data.csv: 01_download.R makefile
$(R_CMD) 01_download.R
02_munged_data.Rds: 02_munge.R 01_downloaded_data.csv outliers_to_remove.txt
$(R_CMD) 02_munge.R
03_model_output.Rds: 03_model.R 02_munged_data.Rds model_config.txt
$(R_CMD) 03_model.R
# Macros
R_CMD = R CMD BATCH --no-save --no-restore --slave --no-timing
R_SCRIPT = R --no-save --no-restore --no-init-file --no-site-file
SET_R_LIBS = .libPaths(c('~/Documents/R/win-library/3.2', \
'C:/Program Files/R/R-3.2.2/library'));
# Targets
all: 04_report_doc.docx
01_downloaded_data.csv: 01_download.R makefile
$(R_CMD) 01_download.R
02_munged_data.Rds: 02_munge.R 01_downloaded_data.csv outliers_to_remove.txt
$(R_CMD) 02_munge.R
03_model_output.Rds: 03_model.R 02_munged_data.Rds model_config.txt
$(R_CMD) 03_model.R
04_report_doc.docx: 04_report.Rmd 03_model_output.Rds
${R_SCRIPT} -e "${SET_R_LIBS} \
knitr::knit(inp='04_report.Rmd', out='04_report.md')"
pandoc 04_report.md --to docx --output 04_report_doc.docx
clean:
rm -f *.Rout
rm -f 04_report.md

